Master thesis: HyperOrdering
Creation date: 2021-07-31
This is the second post of this short series about my master thesis. In part one I explain the problem and the idea of a solution.
As a very small recap: We want to find a permutation of a sparse matrix such that solving a linear system of equations Ax = b by decomposition does not make the decomposition factors too dense. The idea was to use a graph representation for the sparse matrix. We talked about cliques then and decided to eliminate the node which has minimum degree to reduce the number of edges added by creating a clique.
That short recap can only make sense if you read the first post, but then it should. 😉
In this post we want to have a look at the main part of the implementation of my thesis. Again without showing the code here but instead some pseudocode and animations which should give a good idea of how it works.
Naive Pseudocode
We start with the basic pseudocode for our algorithm:
ordering = []
G = (V, E)
while !isempty(V)
v = argmin(deg(u) for u in V)
form_clique(G, N(v))
remove_edges(G, (v,w) in E)
remove(G, v)
ordering = ordering + v
return orderingThis does not create the permutation matrix it just provides a reordering of the node ids but that is enough. Normally one doesn't really use the permutation matrix directly anyway.
I assume here that we get our graph by having a list of vertices and edges. Now as long as we still have vertices to order we take the vertex with minimum degree. Then we have three steps for updating our graph \(G\):
Creating a clique of the neighbors of our node
discussed in the previous post why this corresponds to the fill-in in our matrix later on
remove the edges that connected \(v\) to the rest
remove the vertex \(v\) itself
Now computing
v = argmin(deg(u) for u in V)has normally a running time of \(\mathcal{O}(n)\) when we assume that deg(u) is stored such that accessing the degree of each node can be done in constant time.
form_clique(G, (v,w) in E)would have a running time of \(\mathcal{O}(n^2)\) as each node can be connected to each other node (if we don't assume sparsity here).
In general this would have a running time of \(\mathcal{O}(n^3)\).
Obtaining the node with minimum degree can be speed up dramatically by using some kind of priority queue.
I explained priority queues a bit before for getting the next node in my constraint solver. There I also have some links to more information. Please feel free to have a look at that post.
The basics are a data structure which holds priorities for each element and allows for the following functions:
insert a new element with a given priority
change the priority of an element
get the element with highest priority
remove the element with highest priority
here "highest" can be "lowest" when we multiply everything with \(-1\).
That way getting the next node is a constant operation we just need to update the degrees with some extra cost later.
The problem of forming cliques however is the bigger problem was we have to check every time whether an edge already exists and add it otherwise.
Speedup using Cliques
Currently we are working with only one representation of our matrix which is something like an adjacency list. In an adjacency list we have a list of neighbors for each node which is allows us to add and remove (depends on the implementation) edges in constant time. However checking whether an edge exists isn't fast as we need to go through the list of neighbors. One can improve the running time here by having a hash list which can check whether the key/edge exists.
However checking whether an edge exists still takes time and it will happen often, that the needed edges to form a clique already exist. The idea now is to save the list of cliques itself in addition to the adjacency list. Please make sure to checkout the theoretical paper about this approach Cummings et al. (2021) after reading this post for more information. My master thesis is heavily based on that paper.
To explain this we need some additional fundamentals:
A complete graph with nodes \(U\) is called \(K_U\)
A union of graphs like \(G_1 \cup G_2\) is defined as follows:
Furthermore we use the word hyperedges as an extension to edges. Hyperedges can have 2 or more nodes they are connected. So \(\{1,2\}\) is an edge from node \(1\) to node \(2\) and \(\{1,2,3\}\) is a hyperedge which represents a clique of those three nodes.
We can store the graph as hyperedges in the following way:
\[\begin{aligned} & U_1, U_2, \dots, U_k \subseteq G \\ \text{such that } & G = K_{U_1} \cup K_{U_2} \cup \dots \cup K_{U_k} \end{aligned}\]This just means that our list of hyperedges completely represent the edges in our graph.
Our goal is now that the new representation helps us to reduce the number of checks whether edges need to be added or exist already. We can get a glimpse of that by having a look at the following: Let \(U_1, \dots, U_t\) be the hyperedges that contain our minimum degree node \(v\). Then the new clique we need to add is: \(W = (U_1 \cup \dots \cup U_t) \setminus \{v\}\) and we only need to add the edges which are not already in \((K_{U_1} \cup \dots \cup K_{U_t})\) because those are already part of \(G\).
This should get more clear when we have a look at a visualization. Let's have a look at some pseudocode for that as well which will be used for the animation.
# Initialize the adjacency lists
# Initialize the hyperedges as the edges of the graph
ordering = []
while !isempty(V)
v = minimum_degree_vertex()
ordering = ordering + v
W = Hyperedge()
for U in hyperedges_contain(v)
remove_from_hyperedges(U)
U = U \ {v}
X = W \ U
Y = U \ W
for (x,y) in (X,Y)
# Add edge {x,y} if it doesn't exist
for w in Y
# Remove edge {v,w}
W = union(W,Y)
# add W to list of hyperedges
V = V \ {v}
return orderingIt is not as straight forward as the pseudocode before but I'll try to explain what is happening here with some explanatory images.
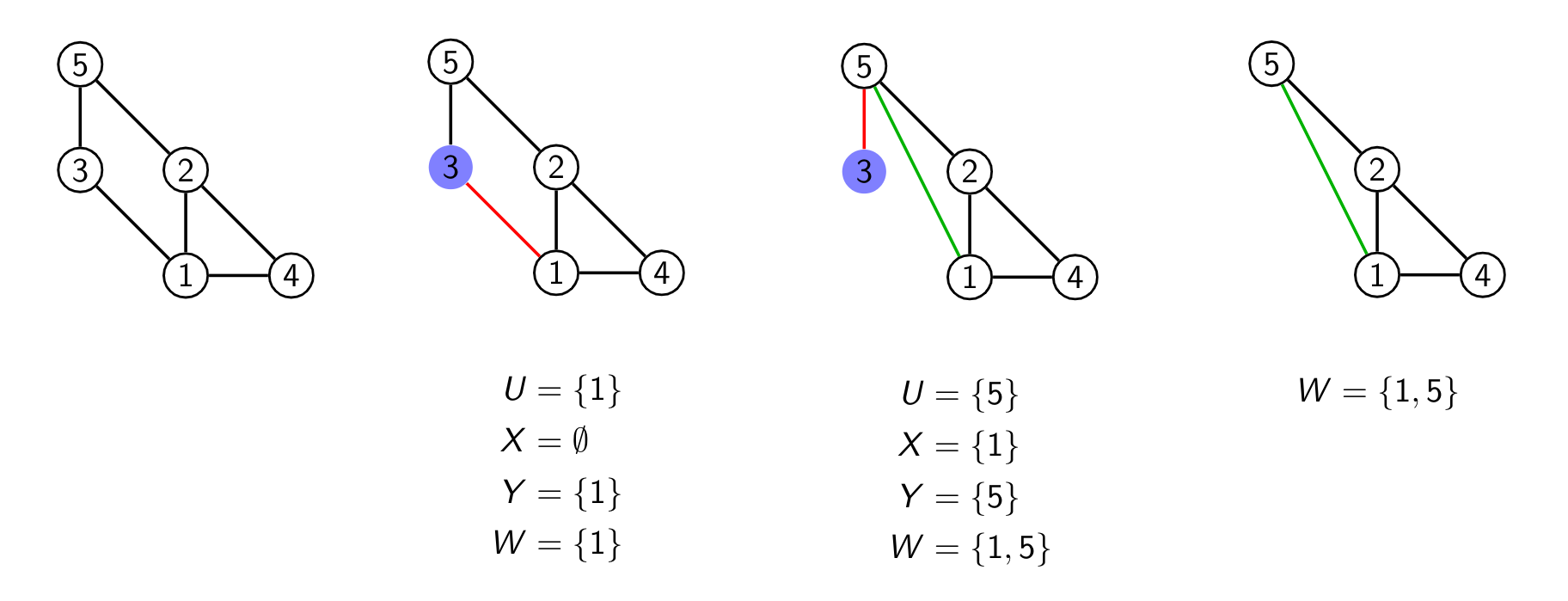
We start with a graph with 5 vertices here and pick vertex 3 as it has only two edges. We could have also picked 4 or 5 here. Now there are two hyperedges which contain the vertex 3 namely {1,3} and {3,5}. I picked again one of them ({1,3}) and it is marked in red as we remove that edge later. We first remove the hyperedge {1,3} from our representation. Then we form U,X,Y which are all written below that second graph in the series above.
In the third step we pick our new hyperedge as we are still iterating over the hyperedges and pick the next one {3,5}. As W isn't empty anymore we do some more work to compute U,X,Y again. This time as both X and Y are not empty we add our new edge connecting 1 and 5. We of course also removed the hyperedge {3,5} from our list and remove the edge {3,5}.
In the last part of the image you can see the additional edge and the hyperedge presentation we added.
Conclusion
This part was a bit shorter but hopefully gives you a broad idea of how the idea from last time can be implemented in a fast way. Please check out the original paper Cummings et al. (2021) if you want way more detail. It covers the kind data structures you want to use to update the elimination graphs. The running time of this algorithm is \(\mathcal{O(nm)}\) with \(n\) being the number of vertices and \(m\) the number of edges. There is also an even tighter bound explained in the paper. For dense matrices this would result in \(\mathcal{O}(n^3)\) again but we want to work with sparse matrices anyway.
Please reach out to me if you're interested in a second part where I would explain the data reduction rules that we used to speed up the overall process by reducing the graph size. Twitter @opensourcesblog
Bibliography
Cummings et al.: A Fast Minimum Degree Algorithm and Matching Lower Bound
Thanks everyone for reading. Hope you enjoyed it and learned something. If you're repeatably happy on this blog please consider subscribing either on Patreon or the newsletter first 😉
Thanks to my 13 patrons!
Special special thanks to my >4$ patrons. The ones I thought couldn't be found 😄
Anonymous
Kangpyo
Gurvesh Sanghera
Szymon Bęczkowski
Colin Phillips
Jérémie Knuesel
For a donation of a single dollar per month you get early access to these posts. Your support will increase the time I can spend on working on this blog.
There is also a special tier if you want to get some help for your own project. You can checkout my mentoring post if you're interested in that and feel free to write me an E-mail if you have questions: o.kroeger <at> opensourc.es
I'll keep you updated on Twitter OpenSourcES.
Want to be updated? Consider subscribing on Patreon for free


